Mohammed M. Abdelsamea
CAPRI-CT: Causal Analysis and Predictive Reasoning for Image Quality Optimization in Computed Tomography
Jul 23, 2025Abstract:In computed tomography (CT), achieving high image quality while minimizing radiation exposure remains a key clinical challenge. This paper presents CAPRI-CT, a novel causal-aware deep learning framework for Causal Analysis and Predictive Reasoning for Image Quality Optimization in CT imaging. CAPRI-CT integrates image data with acquisition metadata (such as tube voltage, tube current, and contrast agent types) to model the underlying causal relationships that influence image quality. An ensemble of Variational Autoencoders (VAEs) is employed to extract meaningful features and generate causal representations from observational data, including CT images and associated imaging parameters. These input features are fused to predict the Signal-to-Noise Ratio (SNR) and support counterfactual inference, enabling what-if simulations, such as changes in contrast agents (types and concentrations) or scan parameters. CAPRI-CT is trained and validated using an ensemble learning approach, achieving strong predictive performance. By facilitating both prediction and interpretability, CAPRI-CT provides actionable insights that could help radiologists and technicians design more efficient CT protocols without repeated physical scans. The source code and dataset are publicly available at https://github.com/SnehaGeorge22/capri-ct.
CLOG-CD: Curriculum Learning based on Oscillating Granularity of Class Decomposed Medical Image Classification
May 03, 2025



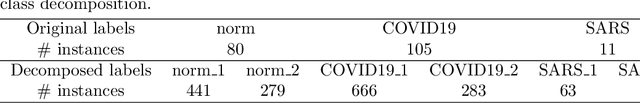
Abstract:Curriculum learning strategies have been proven to be effective in various applications and have gained significant interest in the field of machine learning. It has the ability to improve the final model's performance and accelerate the training process. However, in the medical imaging domain, data irregularities can make the recognition task more challenging and usually result in misclassification between the different classes in the dataset. Class-decomposition approaches have shown promising results in solving such a problem by learning the boundaries within the classes of the data set. In this paper, we present a novel convolutional neural network (CNN) training method based on the curriculum learning strategy and the class decomposition approach, which we call CLOG-CD, to improve the performance of medical image classification. We evaluated our method on four different imbalanced medical image datasets, such as Chest X-ray (CXR), brain tumour, digital knee X-ray, and histopathology colorectal cancer (CRC). CLOG-CD utilises the learnt weights from the decomposition granularity of the classes, and the training is accomplished from descending to ascending order (i.e., anti-curriculum technique). We also investigated the classification performance of our proposed method based on different acceleration factors and pace function curricula. We used two pre-trained networks, ResNet-50 and DenseNet-121, as the backbone for CLOG-CD. The results with ResNet-50 show that CLOG-CD has the ability to improve classification performance with an accuracy of 96.08% for the CXR dataset, 96.91% for the brain tumour dataset, 79.76% for the digital knee X-ray, and 99.17% for the CRC dataset, compared to other training strategies. In addition, with DenseNet-121, CLOG-CD has achieved 94.86%, 94.63%, 76.19%, and 99.45% for CXR, brain tumour, digital knee X-ray, and CRC datasets, respectively
MCUa: Multi-level Context and Uncertainty aware Dynamic Deep Ensemble for Breast Cancer Histology Image Classification
Aug 24, 2021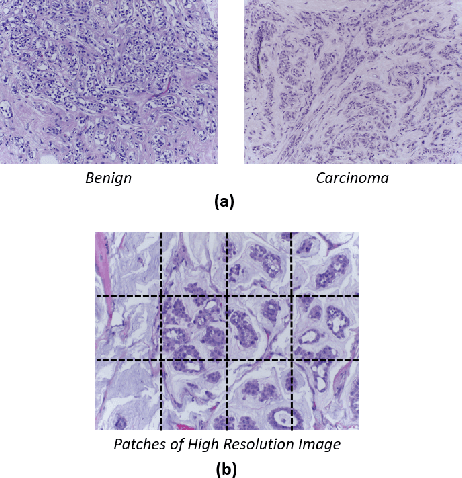
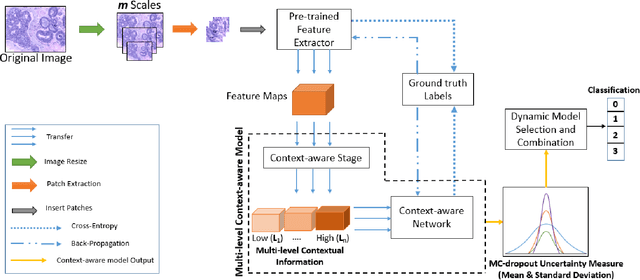
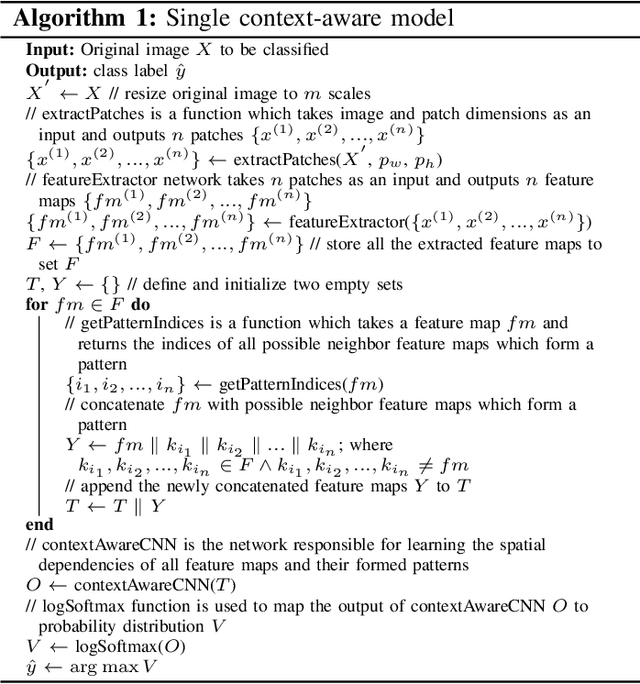
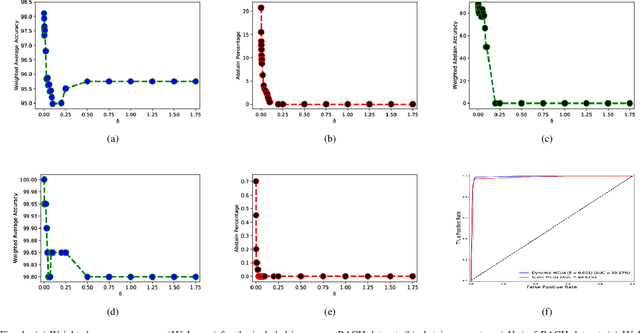
Abstract:Breast histology image classification is a crucial step in the early diagnosis of breast cancer. In breast pathological diagnosis, Convolutional Neural Networks (CNNs) have demonstrated great success using digitized histology slides. However, tissue classification is still challenging due to the high visual variability of the large-sized digitized samples and the lack of contextual information. In this paper, we propose a novel CNN, called Multi-level Context and Uncertainty aware (MCUa) dynamic deep learning ensemble model.MCUamodel consists of several multi-level context-aware models to learn the spatial dependency between image patches in a layer-wise fashion. It exploits the high sensitivity to the multi-level contextual information using an uncertainty quantification component to accomplish a novel dynamic ensemble model.MCUamodelhas achieved a high accuracy of 98.11% on a breast cancer histology image dataset. Experimental results show the superior effectiveness of the proposed solution compared to the state-of-the-art histology classification models.
* accepted by IEEE Transactions on Biomedical Engineering
Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network
Apr 18, 2020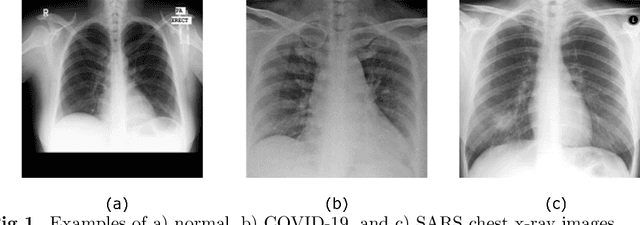
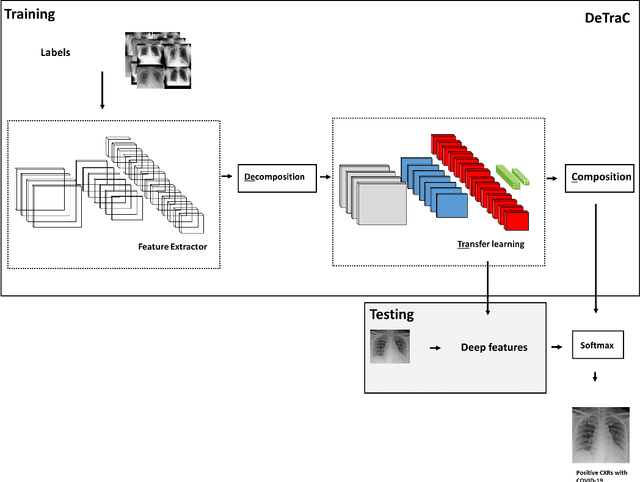
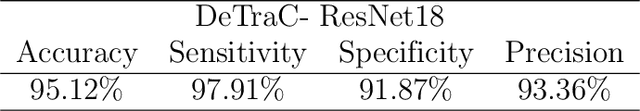
Abstract:Chest X-ray is the first imaging technique that plays an important role in the diagnosis of COVID-19 disease. Due to the high availability of large-scale annotated image datasets, great success has been achieved using convolutional neural networks (CNNs) for image recognition and classification. However, due to the limited availability of annotated medical images, the classification of medical images remains the biggest challenge in medical diagnosis. Thanks to transfer learning, an effective mechanism that can provide a promising solution by transferring knowledge from generic object recognition tasks to domain-specific tasks. In this paper, we validate and adapt our previously developed CNN, called Decompose, Transfer, and Compose (DeTraC), for the classification of COVID-19 chest X-ray images. DeTraC can deal with any irregularities in the image dataset by investigating its class boundaries using a class decomposition mechanism. The experimental results showed the capability of DeTraC in the detection of COVID-19 cases from a comprehensive image dataset collected from several hospitals around the world. High accuracy of 95.12% (with a sensitivity of 97.91%, a specificity of 91.87%, and a precision of 93.36%) was achieved by DeTraC in the detection of COVID-19 X-ray images from normal, and severe acute respiratory syndrome cases.
An Automatic Seeded Region Growing for 2D Biomedical Image Segmentation
Dec 12, 2014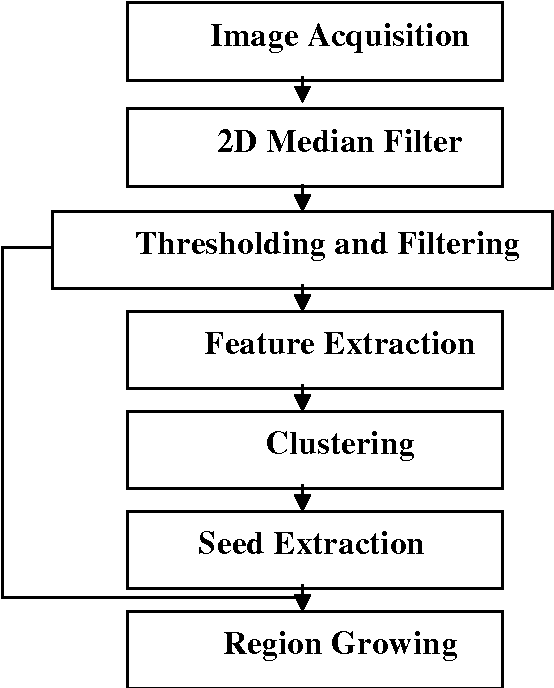
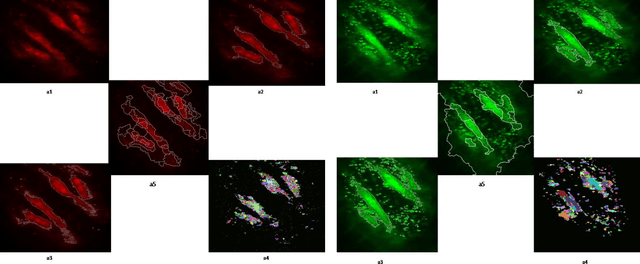
Abstract:In this paper, an automatic seeded region growing algorithm is proposed for cellular image segmentation. First, the regions of interest (ROIs) extracted from the preprocessed image. Second, the initial seeds are automatically selected based on ROIs extracted from the image. Third, the most reprehensive seeds are selected using a machine learning algorithm. Finally, the cellular image is segmented into regions where each region corresponds to a seed. The aim of the proposed is to automatically extract the Region of Interests (ROI) from the cellular images in terms of overcoming the explosion, under segmentation and over segmentation problems. Experimental results show that the proposed algorithm can improve the segmented image and the segmented results are less noisy as compared to some existing algorithms.
Unsupervised Parallel Extraction based Texture for Efficient Image Representation
Aug 20, 2014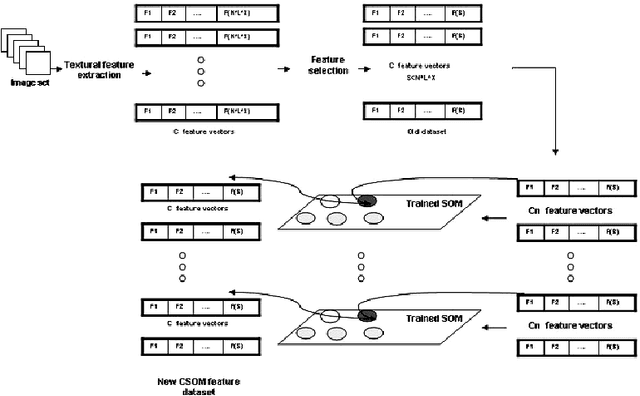
Abstract:SOM is a type of unsupervised learning where the goal is to discover some underlying structure of the data. In this paper, a new extraction method based on the main idea of Concurrent Self-Organizing Maps (CSOM), representing a winner-takes-all collection of small SOM networks is proposed. Each SOM of the system is trained individually to provide best results for one class only. The experiments confirm that the proposed features based CSOM is capable to represent image content better than extracted features based on a single big SOM and these proposed features improve the final decision of the CAD. Experiments held on Mammographic Image Analysis Society (MIAS) dataset.
* arXiv admin note: substantial text overlap with arXiv:1408.4143
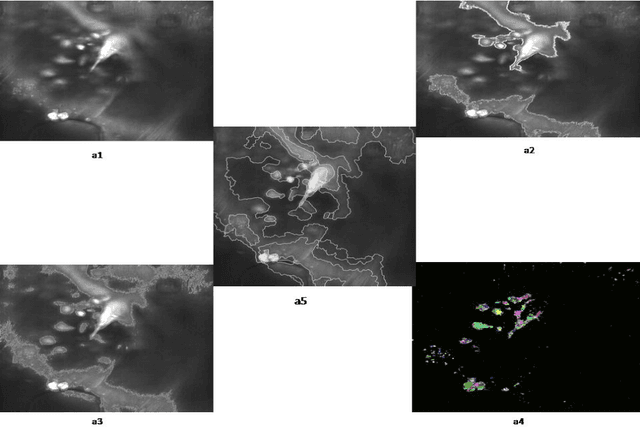
An Enhancement Neighborhood connected Segmentation for 2D-Cellular Image
Jul 14, 2014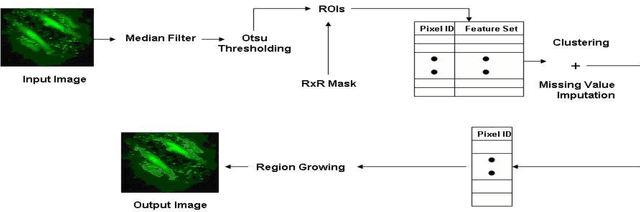
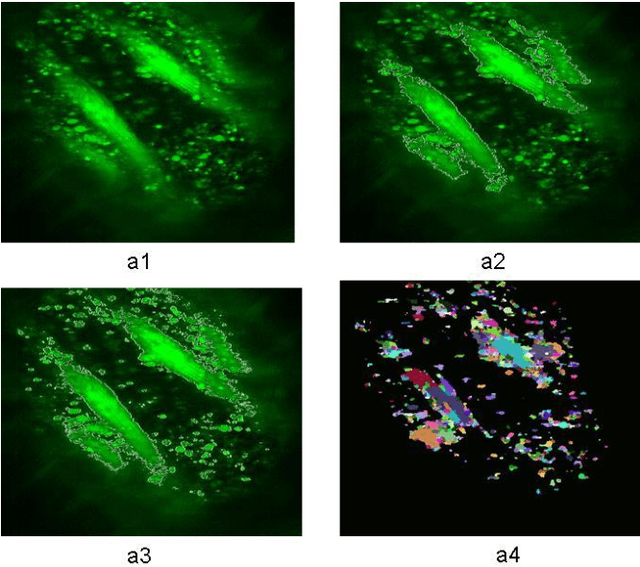
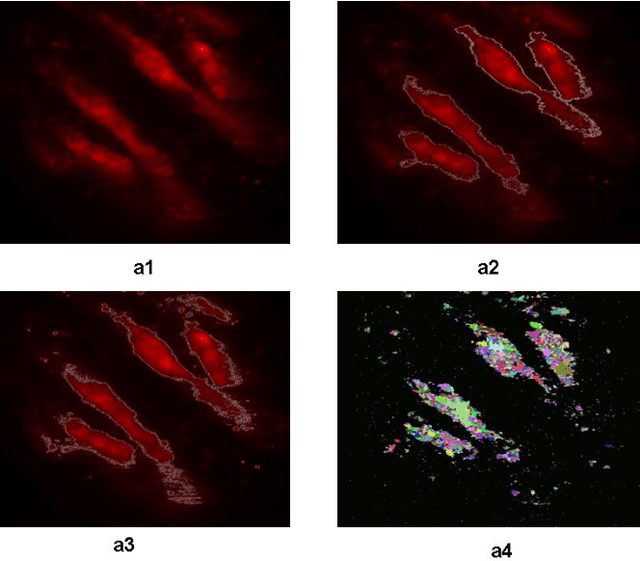
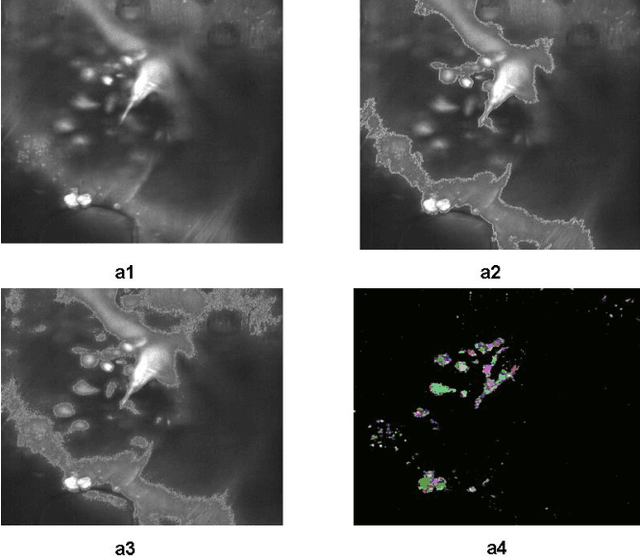
Abstract:A good segmentation result depends on a set of "correct" choice for the seeds. When the input images are noisy, the seeds may fall on atypical pixels that are not representative of the region statistics. This can lead to erroneous segmentation results. In this paper, an automatic seeded region growing algorithm is proposed for cellular image segmentation. First, the regions of interest (ROIs) extracted from the preprocessed image. Second, the initial seeds are automatically selected based on ROIs extracted from the image. Third, the most reprehensive seeds are selected using a machine learning algorithm. Finally, the cellular image is segmented into regions where each region corresponds to a seed. The aim of the proposed is to automatically extract the Region of Interests (ROI) from in the cellular images in terms of overcoming the explosion, under segmentation and over segmentation problems. Experimental results show that the proposed algorithm can improve the segmented image and the segmented results are less noisy as compared to some existing algorithms.
Self Organization Map based Texture Feature Extraction for Efficient Medical Image Categorization
Jul 14, 2014Abstract:Texture is one of the most important properties of visual surface that helps in discriminating one object from another or an object from background. The self-organizing map (SOM) is an excellent tool in exploratory phase of data mining. It projects its input space on prototypes of a low-dimensional regular grid that can be effectively utilized to visualize and explore properties of the data. This paper proposes an enhancement extraction method for accurate extracting features for efficient image representation it based on SOM neural network. In this approach, we apply three different partitioning approaches as a region of interested (ROI) selection methods for extracting different accurate textural features from medical image as a primary step of our extraction method. Fisherfaces feature selection is used, for selecting discriminated features form extracted textural features. Experimental result showed the high accuracy of medical image categorization with our proposed extraction method. Experiments held on Mammographic Image Analysis Society (MIAS) dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge